NumPy: MKL vs OpenBLAS
(Code available at harningle/useful-scripts)
TL;DR: There can be some low hanging performance gains from MKL vs OpenBLAS
It’s a nightmare to install geospatial packages for Python on Windows. And the perfect go-to solution I saw from many answers in Stack Overflow is often Christoph Gohlke’s wheels. He also maintains wheels for NumPy. However, he says these wheels are “linked to the Intel(r) oneAPI Math Kernel Library (oneAPI MKL).” This was the first time I’d heard about stuff like MKL and OpenBLAS. More recently I noticed some performance difference between “usual” NumPy and NumPy built against MKL. Here are how to install NumPy with different BLAS and some benchmark results.
What are they and how to install them?
NumPy depends on some linear algebra library. That “some library” can be Intel MKL (math kernel library), or OpenBLAS (open basic linear algebra subprograms).1 They are different in size, performance, license/whether open souce, and so on. One important feature is, they both usually use all cores for some functions, such as np.dot.
OpenBLAS
OpenBLAS is the default library when we install NumPy in the “usual” way:
conda install numpy -c conda-forge
# Or
# pip install numpy
After installation, you should be able to see something like name: blas or name: scipy-openblas in the NumPy config.:
>>> import numpy as np
>>> np.show_config()
Build Dependencies:
blas:
detection method: pkgconfig
found: true
include directory: /home/wang/miniconda3/envs/np-openblas/include
lib directory: /home/wang/miniconda3/envs/np-openblas/lib
name: blas
openblas configuration: unknown
pc file directory: /home/wang/miniconda3/envs/np-openblas/lib/pkgconfig
version: 3.9.0
lapack:
detection method: pkgconfig
found: true
include directory: /home/wang/miniconda3/envs/np-openblas/include
lib directory: /home/wang/miniconda3/envs/np-openblas/lib
name: lapack
openblas configuration: unknown
pc file directory: /home/wang/miniconda3/envs/np-openblas/lib/pkgconfig
version: 3.9.0
[rest of the output omitted]
You may wonder, hmmm, it says openblas configuration: unknown. Are we really using OpenBLAS? First, NumPy says “[t]he NumPy wheels on PyPI, which is what pip installs, are built with OpenBLAS”, and “[w]hen a user installs NumPy from conda-forge, that BLAS package then gets installed together with the actual library - this defaults to OpenBLAS, but it can also be MKL (from the defaults channel)[…]” Since we specify conda-forge when installing it, there shouldn’t be any problem. If you look at the details when we conda install, it does install the correct OpenBLAS library.
[output above omitted]
The following NEW packages will be INSTALLED:
libcblas conda-forge/linux-64::libcblas-3.9.0-25_linux64_openblas
liblapack conda-forge/linux-64::liblapack-3.9.0-25_linux64_openblas
libstdcxx conda-forge/linux-64::libstdcxx-14.2.0-hc0a3c3a_1
numpy conda-forge/linux-64::numpy-2.1.3-py312h58c1407_0
[the rest of output omitted]
We can also go further into the dependencies of NumPy.2
$ ldd <PATH-TO-YOUR-PYTHON>/site-packages/numpy/_core/_multiarray_umath.cpython-312-x86_64-linux-gnu.so
linux-vdso.so.1 (0x00007ffca5249000)
libcblas.so.3 => [omitted]/libcblas.so.3 (0x00007fee4e696000)
libstdc++.so.6 => [omitted]/libstdc++.so.6 (0x00007fee51156000)
libm.so.6 => /lib64/libm.so.6 (0x00007fee4e314000)
libgcc_s.so.1 => [omitted]/libgcc_s.so.1 (0x00007fee4e2f5000)
libc.so.6 => /lib64/libc.so.6 (0x00007fee4df1f000)
/lib64/ld-linux-x86-64.so.2 (0x00007fee5110b000)
libpthread.so.0 => /lib64/libpthread.so.0 (0x00007fee4dcff000)
libgfortran.so.5 => [omitted]/libgfortran.so.5 (0x00007fee4db5a000)
libdl.so.2 => /lib64/libdl.so.2 (0x00007fee4d956000)
librt.so.1 => /lib64/librt.so.1 (0x00007fee4d74e000)
libquadmath.so.0 => [omitted]/libquadmath.so.0 (0x00007fee4d716000)
$ readlink -e [omitted]/libcblas.so.3
[omitted]/libopenblasp-r0.3.28.so
So eventually NumPy is using libopenblasp-r0.3.28.so. No problem.
MKL
MKL is from Intel. It may have some problem with commercial use or whatever. Despite coming from Intel, you can use it on AMD CPU or Apple silicon. Installing NumPy with MKL is slightly more complicated than OpenBLAS. If we use pip, I find it most easily to use Christoph Gohlke’s NumPy wheels. For example, to install his v2024.10.14 wheel on Python 3.10, we run:
pip install https://github.com/cgohlke/numpy-mkl-wheels/releases/download/v2024.10.14/mkl_service-2.4.1-cp310-cp310-win_amd64.whl https://github.com/cgohlke/numpy-mkl-wheels/releases/download/v2024.10.14/numpy-2.1.2-cp310-cp310-win_amd64.whl
Conda environment is easier on this occasion:
conda install numpy libblas=*=*mkl -c conda-forge
Depending on which conda channel we are using, we may or may not need to manually specify libblas=*=*mkl (but it’s always safer to specify so). Check conda-forge docs for details about switching BLAS implementation.
Again, after installing NumPy in this way, we should see things like name: mkl-sdl in NumPy config.:
>>> import numpy as np
>>> np.show_config()
Build Dependencies:
blas:
detection method: pkgconfig
found: true
include directory: /home/wangxu/miniconda3/envs/np-mkl/include
lib directory: /home/wangxu/miniconda3/envs/np-mkl/lib
name: mkl-sdl
openblas configuration: unknown
pc file directory: /home/wangxu/miniconda3/envs/np-mkl/lib/pkgconfig
version: '2023.1'
lapack:
detection method: pkgconfig
found: true
include directory: /home/wangxu/miniconda3/envs/np-mkl/include
lib directory: /home/wangxu/miniconda3/envs/np-mkl/lib
name: mkl-sdl
openblas configuration: unknown
pc file directory: /home/wangxu/miniconda3/envs/np-mkl/lib/pkgconfig
version: '2023.1'
[rest of the output omitted]
Benchmark
I run several frequently used functions to benchmark the performance for both BLAS: matrix multiplication, inverse, eigenvalue, and decomposition. When NumPy is working hard, we do see all cores are being utilised. In my use case, this is often a good thing: I don’t do fancy multi threading on my own, so it’s better if NumPy automatically squeezes all my computer’s power.3
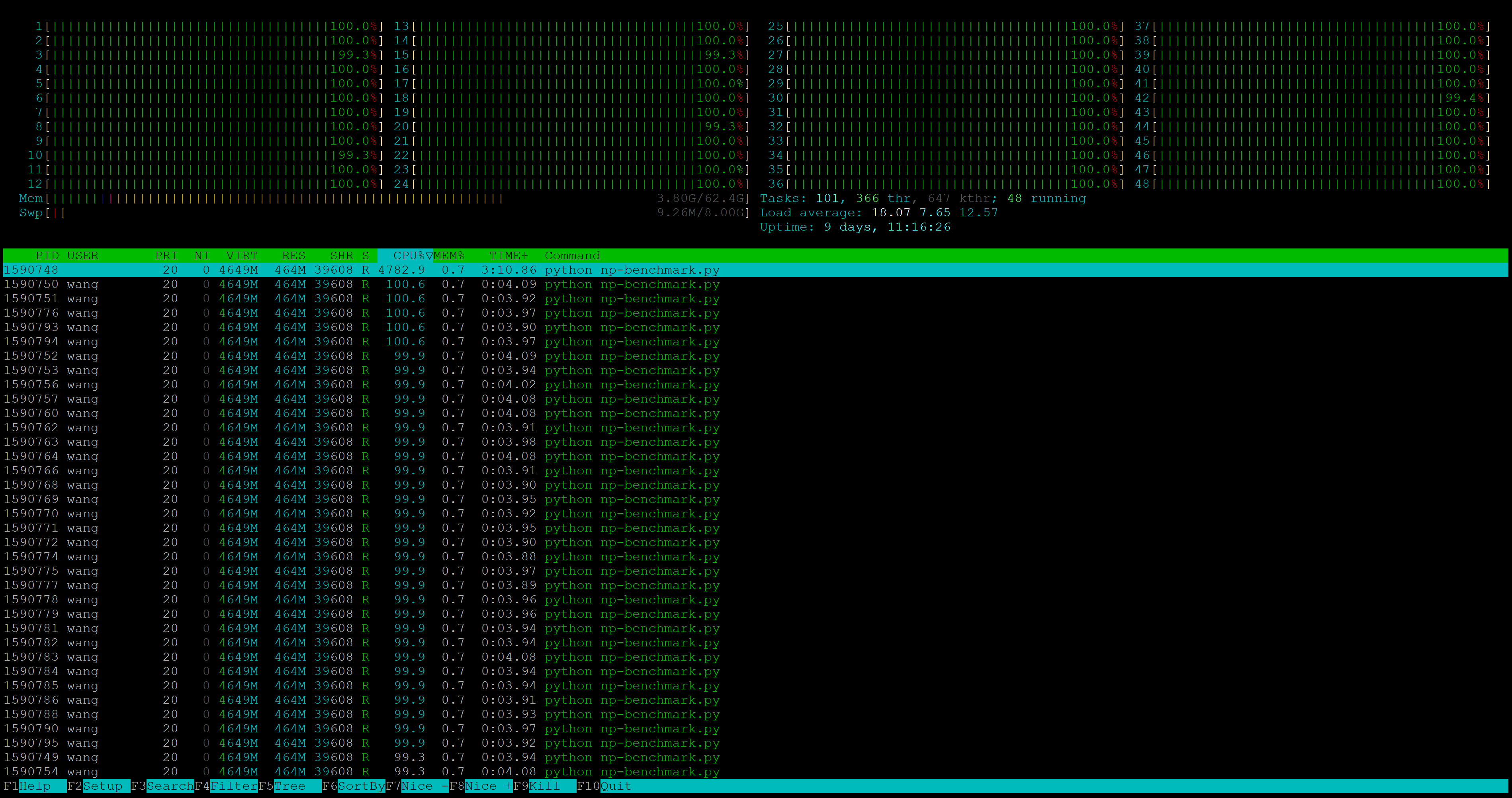
Notes: The benchmark was run on a 24-core Intel Xeon Gold 6248R CPU.
The full benchmark results are shown below. MKL is much faster across the board, with vector norm being the only exception (which I don’t know why). This result can vary across machines. On my personal desktop with a 6-core Intel middle-low tier consumer CPU, the results are very much tied. It’s worth running a few benchmarks if you really care about the speed. At some superficial level, MKL’s speed over OpenBLAS seems a free lunch to me.
Notes: The speed up factor is calculated as MKL time over OpenBLAS time, so a factor of 0.1 means MKL takes only 10% of the time required by OpenBLAS to perform the same operation. The benchmark was run on a 24-core Intel Xeon Gold 6248R CPU. The vector being used is a $5000\times 1$ float64 column vector, and the matrix is also float 64, has a size of $5000\times 5000$, and is positive semi-definite. The time is averaged across ten runs.
-
We do have other BLAS packages available, such as BLIS. See NumPy docs. ↩
-
See https://stackoverflow.com/questions/37184618/find-out-if-which-blas-library-is-used-by-numpy for details. ↩
-
If you are running your own multiprocessing, e.g. using
multiprocessor Dask, NumPy’s multiprocessing can be a problem. To disable it, setos.environ['OPENBLAS_NUM_THREADS'] = '1'oros.environ['MKL_NUM_THREADS'] = '1'. ↩